Nucleic Acids
Category : 12th Class
Two types of nucleic acids are found in the cells of all living organisms. These are DNA (Deoxyribonucleic acid) and RNA (Ribonucleic acid). The nucleic acid was first isolated (reported) by Friedrich Miescher in 1869 from the nuclei of pus cells and was named nuclein. The term nucleic acid was given by Altman (1899).
![]()
Term was given by Zacharis, which is found in the cells of all living organisms except plant viruses, where RNA forms the genetic material and DNA is absent. In bacteriophages and viruses there is a single molecule of DNA, which remains coiled and is enclosed in the protein coat. In bacteria, mitochondria, plastids and other prokaryotes, DNA is circular and lies naked in the cytoplasm but in eukaryotes it is found in nucleus and known as carrier of genetic information and capable of self replication. Isolation and purification of specific DNA segment from a living organism achieved by Nirenberg. H.Harries is associated with DNA-RNA hybridization technique.
Chemical composition
The chemical analysis has shown that DNA is composed of three different types of compound.
(i) Sugar molecule : Levene identified a five carbon sugar, ribose in nucleic acid in 1910. It is represented by a pentose sugar the deoxyribose or 2-deoxyribose which derived from ribose due to the deletion of oxygen from the second carbon.
(ii) Phosphoric acid : \[{{H}_{3}}P{{O}_{4}}\] that makes DNA acidic in nature.
(iii) Nitrogeneous base : Kossel demonstrated the presence of two pyrimidines (cytosine and thymine) and two purines (adenine and guanine) in DNA and he was awarded Nobel Prize in 1910. These are nitrogen containing ring compound. Which classified into two groups:
(a) Purines : Two ring compound namely as Adenine and Guanine.
(b) Pyrimidine : One ring compound included Cytosine and Thymine. In RNA Uracil is present instead of Thymine.
Nucleosides : Nucleosides are formed by a purine or pyrimidine nitrogenous base and pentose sugar. DNA nucleosides are known as deoxyribosenucleosides.
Nucleotides : In a nucleotide, purine or pyrimidine nitrogenous base is joined by deoxyribose pentose sugar (D), which is further linked with phosphate (P) group to form nucleotides.
|
Nitrogenous base |
Nucleoside (Base + Sugar) |
Nucleotide (Base + Sugar + Phosphate) |
|
DNA Adenine = A |
Deoxyadenosine |
Deoxyadenosine monophosphate or Adenine deoxyribose nucleotide |
|
Guanine = G |
Deoxyguanine |
Deoxygunine monophosphate or Guanine deoxyribose-nucleotide |
|
Thyamine = T |
Thymidine |
Deoxythymidine monophosphate or Thymidine deoxyribose nucleotide |
|
Cytosine = C |
Deoxycitidine |
Deoxycytidine monophosphate or Cytosine deoxyribose nucleotide |
|
RNA Adenine = A |
Adenosine |
Adenosine monophosphate or Adenine ribose nucleotide |
|
Guanine = G |
Guanosine |
Guanosine monophosphate or Guanine ribose nucleotide |
|
Uracil = U |
Uridine |
Uridine monophosphate or Uracil ribose nucleotide |
|
Cytosine = C |
Cytidine |
Cytidine monophosphate or Cytosine ribose nucleotide |
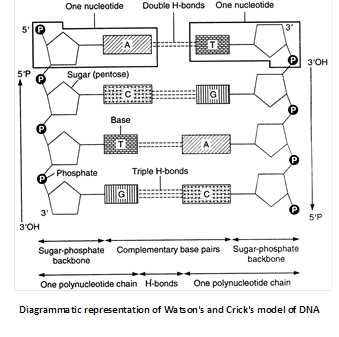
Watson and Crick’s model of DNA
In 1953 James Watson and Francis Crick suggested that in a DNA molecule there are two such polynucleotide chains arranged antiparallal or in opposite directions i.e., one polynucleotide chain runs in \[{{5}^{}}\to {{3}^{}}\] direction, the other in \[{{3}^{}}\to {{5}^{}}\] direction. It means the 3’ end of one chain lies beside the 5’ end of other in right handed manner.
Important features
(i) The double helix comprises of two polynucleotide chains.
(ii) The two strands (polynucleotide chains) of double helix are anti-parallel due to phosphodiester bond.
(iii) Each polynucleotide chain has a sugar-phosphate ‘backbone’ with nitrogeneous bases directed inside the helix.
(iv) The nitrogenous bases of two antiparallel polynucleotide strands are linked through hydrogen bonds. There are two hydrogen bonds between A and T, and three between G and C. The hydrogen bonds are the only attractive forces between the two polynucleotides of double helix. These serve to hold the structure together.
The two polynucleotides in a double helix are complementary. The sequence of nitrogenous bases in one determines the sequence of the nitrogenous bases in the other. Complementary base pairing is of fundamental importance in molecular genetics.
Erwin Chargaff (1950) made quantitative analysis of DNA and proposed “base equivalence rule” starting that molar concentration of \[A=T\And G\equiv C\]or \[\frac{A+G}{C+T}=1\And \frac{A+T}{G+C}\]which is constant for a species. Sugar deoxyribose and phosphate occur in equimolar proportion.
Ten base pairs occur per turn of helix (abbreviated 10bp). The spacing between adjacent base pairs is \[3.4{AA}.\] The helix is \[20{AA}\text{ (}19.8{AA}\text{)}\]in diameter and DNA molecule found 360o in a clockwise.
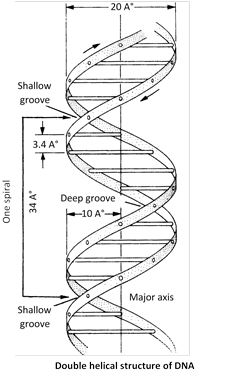
Forms of DNA
Five different morphological forms of DNA double helix have been described. These are A, B, C, D and Z forms. Most of these forms (except B, and Z) occur in rigidly controlled experimental conditions. Watson and crick model represents commonest form, Biotic-form (B-form or B-DNA) of DNA. Some DNA forms are inter convertible also. The differences in these DNA forms are associated with :
(i) The numbers of base pairs, present in each turn of DNA helix.
(ii) The pitch or angle between each base pair.
(iii) The helical diameter of DNA molecule.
(iv) The handedness of double helix. Which is mentioned in table.
Comparison of different types of DNA
|
Characters |
A-DNA |
B-DNA |
C-DNA |
D-DNA |
Z-DNA |
|
Base pair per turn of the helix |
11 |
10 |
9.33 |
8 |
12 |
|
Tilt of pairs (\[\gamma \]) base |
20.20 |
6.30 |
-7.80 |
-16.70 |
7 Å |
|
Axial rise (h) |
2.56 Å |
3.37 Å |
3.32 Å |
3.03 Å |
3.7 Å |
|
Helical diameter |
23 Å |
20 Å |
19 Å |
- |
18 Å |
|
Handedness of the double helix |
Right handed |
Right handed |
Right handed |
Right handed |
Left handed |
Promiscuous DNA : Special type of DNA which makes movement between mitochondria, chloroplast and nucleus. It was discovered in 1983 in cambridge university in maize. It was later reported in yeast, mungbean, spinach and peas.
Repetitive DNA : Multiple copies of DNA having same or almost same base pair sequence constitute repetitive DNA. In higher organisms \[20%90%\] DNA is of this type.
Satellite DNA : In some eukaryotes small highly repetitive DNA sequences have been found called satellite DNA, which differ in base composition.
Characteristics of DNA
Denaturation or melting : The phenomenon of separating of two strand of DNA molecule by breaking of hydrogen bond at the temp. \[{{90}^{{}^\circ }}C.\] In prokaryotes and human mitochondria \[G\equiv C\] are more because this melting point is more. In eukaryotes the amount of \[A=T\]are more because melting point is less.
Renaturation or annealing : Separated strands reunite to form double helix molecule of DNA by cooling at the room temp. i.e., \[25{}^\circ C.\]
These properties help to form hybrid from different DNA or with RNA.
Evidences of DNA as the genetic material
The following experiments conducted by the molecular biologists provide direct evidences of DNA being the genetic material.
Bacterial transformation or Griffith's Experiments : Griffith (1928) injected into mice with virulent and smooth (S-type, smooth colony with mucilage) form of Diplococcus pneumoniae. The mice died due to pneumonia. No death occurred when mice were injected with nonvirulent or rough (R-type, irregular colony without mucilage) form or heat- killed virulent form. However, in a combination of heat killed S-type and live R-type bacteria, death occurred in some mice. Autopsy of dead mice showed that they possessed S-type living bacteria, which could have been produced only by transformation of R-type bacteria. The transforming chemical was found out by O.T. Avery, C.M. Mc. leod and M. Mc. Carty (1944). They fractionated heat-killed S-type bacteria into DNA, carbohydrate and protein fractions. DNA was divided into two parts, one with DNAase and the other without it. Each component was added to different cultures of R-type bacteria. Transformation was found only in that culture which was provided with intact DNA of S-type. Therefore, the trait of virulence is present in DNA. Transformation involves transfer of a part of DNA from surrounding medium or dead bacteria (donor) to living bacteria (recipient) to form a recombinant.
Evidence from genetic recombination in bacteria or bacterial conjugation : Lederberg and Tatum (1946) discovered the genetic recombination in bacteria from two different strains through the process of conjugation. Bacterium Escherichia coli can grow in minimal culture medium containing minerals and sugar only. It can synthesize all the necessary vitamins from these raw materials. But its two mutant strains were found to lack the ability to synthesize some of the vitamins necessary for growth. These could not grow in the minimal medium till the particular vitamins were not supplied in the culture medium.
Mutant strain A : It (used as male strain) had the genetic composition \[Me{{t}^{}},Bi{{o}^{}},Th{{r}^{+}},Le{{u}^{+}},Th{{i}^{+}}.\] It lacks the ability to manufacture vitamins methionine and biotin and can grow only in a culture medium which contains these vitamins in addition to sugar and minerals.
Mutant strain B : It (used as female strain or recipient) has a genetic composition \[M{{e}^{++}},Bi{{o}^{+}},Th{{r}^{}},Le{{u}^{}},~Th{{i}^{}}.\]It lacks the ability to manufacture threonine, leucine and thionine and can grow only when these vitamins are added to the growing medium.
These two strains of E.coli are, therefore, unable to grow in the minimal culture medium, when grow separately. But when a mixture of these two strains was allowed to grow in the same medium a number of colonies were formed. This indicates that the portion of donor DNA containing information to manufacture threonine, leucine and thionine had been transferred and incorporated in the recipient’s genotype during conjugation.
This experiment of Lederberg and Tatum shown that the conjugation results in the transfer of genetic material DNA from one bacterium to other. During conjugation a cytoplasmic bridge is formed between two conjugating bacteria.
Evidence from bacteriophage infection : Hershey and Chase (1952) conducted their experiment on \[{{T}_{2}}\]bacteriophage, which attacks on E.coli bacterium. The phage particles were prepared by using radioisotopes of \[{{S}^{35}}\]and \[{{P}^{32}}\]in the following steps :
(i) Few bacteriophages were grown in bacteria containing \[^{35}S.\] Which was incorporated into the cystein and methionine amino acids of proteins and thus these amino acids with \[^{35}S\] formed the proteins of phage.
(ii) Some other bacteriophages were grown in bacteria having 32P. Which was restricted to DNA of phage particles. These two radioactive phage preparations (one with radioactive proteins and another with radioactive DNA) were allowed to infect the culture of E.coli. The protein coats were separated from the bacterial cell walls by shaking and centrifugation.
The heavier infected bacterial cells during centrifugation pelleted to bottom. The supernatant had the lighter phage particles and other components that failed to infect bacteria. It was observed that bacteriophages with radioactive DNA gave rise to radioactive pellets with \[^{32}P\] in DNA. However in the phage particles with radioactive protein (with\[^{35}S\]) the bacterial pellets have almost nil radioactivity indicating that proteins have failed to migrate into bacterial cell. So, it can be safely concluded that during infection by bacteriophage T2, it was DNA, which entered the bacteria. It was followed by an eclipse period during which phage DNA replicates numerous times within the bacterial cell. Towards the end of eclipse period phage DNA directs the production of protein coats assembly of newly formed phage particles. Lysozyme (an enzyme) brings about the lysis of host cell and release, the newly formed bacteriophages. The above experiment clearly suggests that it is phage DNA and not protein, which contains the genetic information for the production of new bacteriophages. However, in some plant viruses (like TMV), RNA acts as hereditary material (being DNA absent).
DNA replication
Watson and Crick suggested a very simple mechanism of DNA replication or DNA transcription on the basis of its double helical structure. During replication the weak hydrogen bonds between the nitrogeneous bases of the nucleotides separate so that the two polynucleotide chains of DNA also separate and uncoil. The chains thus separated are complementary to one another. Because of the specificity of base pairing, each nucleotide of separated chains attracts it complementary nucleotide from the cell cytoplasm. Once the nucleotides are attached by their hydrogen bonds, their sugar radicals unite through their phosphate components, completing the formation of a new polynucleotide chain.
The method of DNA replication is semi-discontinuous and described as semi-conservative method, because each daughter DNA molecule is a hybrid conserving one parental polynucleotide chain and the other one newly synthesized strand. DNA replication occur in S-phage in cell cycle.
Mechanism of DNA replication
The entire process of DNA replication involves following steps in E.coli :
Recognition of the initiation point : First, DNA helix unwind by the enzyme “Helicase” which use the energy of ATP and replication of DNA begin at a specific point, called initiation point or origin where replication fork begins.
Unwinding of DNA : The unwinding proteins bind to the nicked strand of the duplex and separate the two strands at DNA duplex. Topoisomerase (Gyrase is a type of topoisomerase in E.coli) helps in unwinding of DNA.
Single stranded binding protein (SSB) : Which remain DNA in single stranded position and also known as helix destabilising protein (HDP).
RNA Priming : The DNA directed RNA polymerase now synthesizes the primer strands of RNA (RNA primer). The priming RNA strands are complementary to the two strands of DNA and are formed of 50 to 100 nucleotides.
Formation of DNA on RNA primers : The new strands of DNA are formed in the \[{{5}^{'}}\to {{3}^{'}}\] direction from the \[{{3}^{'}}\to {{5}^{'}}\] template DNA by the addition of deoxyribonucleotides to the \[{{3}^{'}}\]end of primer RNA.
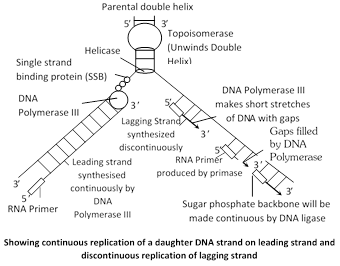
Addition of nucleotide is done by DNA polymerase III. The leading strand of DNA is synthesized continuously in \[{{5}^{'}}\to {{3}^{'}}\] direction as one piece. The lagging strand of DNA is synthesized discontinuously in its opposite direction in short segments. These segments are called Okazaki fragments.
Excision of RNA primers : Once a small segment of an okazaki fragment has been formed. The RNA primers are removed from the 5¢ by the action of \[{{5}^{'}}\to {{3}^{'}}\]exonuclease activity of DNA polymerase I.
Joining of okazaki fragments : The gaps left between Okazaki fragments are filled with complimentary deoxyribonucleotide residues by DNA polymerase-I. Finally, the adjacent 5¢ and 3¢ ends are joined by DNA ligase.
DNA polymerase enzymes
There are three DNA polymerase enzymes that participate in the process of DNA replication.
(i) DNA polymerase-I : This enzyme has been studied in E. coli in detail. It possesses a sulphydryl group, single interchain disulphide and one zinc molecule at the active site. DNA polymerase-I was discovered by Kornberg and his colleagues in 1955. It was considered to carry out DNA replication and also participates in the repair and proof reading of DNA by catalyzing the addition of mononucleotide units (the deoxyribonucleotide residues) to the free \[{{3}^{'}}-\]hydroxyl end of DNA chain. A pure DNA polymerase-I can add about 1,000 nucleotide residues per minute per molecule and catalyses \[{{5}^{'}}\to {{3}^{'}}\] exonuclease activity and removes nucleotide residues of primer RNA at 3¢.
(ii) DNA polymerase-II : The biological role of polymerase II is not yet known.
(iii) DNA polymerase-III : This enzyme was discovered by T. Kornberg and M.L. Gefter (1972). It is the most active enzyme and responsible for DNA chain elongation.
DNA repair : When DNA damaged by mutagen, a system is activate to repair damage DNA. Say for example UV light induced thymidine dimers in DNA and repair mechanism of that DNA called photoreactivation. Many enzyme involved in repair mechanism in which endonuclease (Chemical knives) cut the defective part of DNA then gap is filled with DNA polymerase I and finally DNA ligase seals that repaired part.
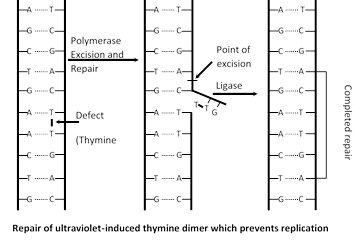
Evidence in support of semiconservative mode of DNA replication (Meselson and Stahl’s experiment)
(1) Meselson and Stahl (1958) cultured (Escherichia coli) bacteria in a culture medium containing \[{{N}^{15}}\] were isotopes of nitrogen. After these had replicated for a few generations in that medium both the strands of their DNA contained \[{{N}^{15}}\] as constituents of purines and pyrimidines. When these bacteria with \[{{N}^{15}}\] were transferred in cultural medium containing \[{{N}^{14}},\]it was found that DNA separated from fresh generation of bacteria possesses one strand heavier than the other. The heavier strand represents the parental strand and lighter one is the new one synthesized from the culture indicating semiconservative mode of DNA replication. circular form of replication on as characteristic of prokaryotes is theta replication discovered by J. Cairns.
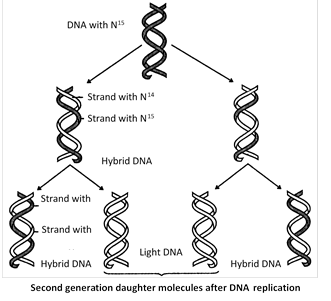
(2) Evidence from Taylor?s experiment on Vicia faba (Broad Bean) root tips using autoradiography technique and further he used tritiated thymidine (H3-tdR).
(3) Evidence from Cairn?s autoradiography experiment in bacteria. He used tritiated thymidine \[({{H}^{3}}-tdR).\]
![]()
RNA is found in the cytoplasm and nucleolus. Inside the cytoplasm it occurs freely as well as in the ribosomes. RNA can also be detected from mitochondria, chloroplasts and associated with the eukaryotic chromosomes. In some plant viruses RNA acts as hereditary material.
Structure of RNA
More commonly RNA is a single stranded structure consisting of an unbranched polynucleotide chain, but it is often folded back on itself forming helices. DNA is a double stranded structure and its two polynucleotide chains are bounded spirally around a main axis. It is made up by :
(1) Sugar : Ribose
(2) Phosphate : In the form of \[{{H}_{3}}P{{O}_{4}}.\]
(3) Nitrogenous base : Two types:
(a) Purine,
(b) Pyramidine
(i) Purine is further divided into Adenine and Guanine.
(ii) Pyramidin divided into Cytosine and Uracil.
Types of RNA
RNA can be classified into two types.
(1) Genetic RNA : Which established by Conrat. In most of the plant viruses, some animal viruses and in many bacteriophages DNA is not found and RNA acts as hereditary material. This RNA may be single stranded or double stranded.
(2) Nongenetic RNA : In the all other organisms where DNA is the hereditary material, different types of RNA are nongenetic. The nongenetic RNA is synthesized from DNA template.
In general, three types of RNAs have been distinguished :
Messenger RNA or Nuclear RNA (mRNA) : mRNA is a polymer of ribo-nucleotide as a complementary strand to DNA and carries genetic information in cytoplasm for the synthesis of proteins. For this reason only, it was named messenger RNA (mRNA) by Jacob and Monod (1961) is 5% of total RNA. It acts as a template for protein synthesis and has a short life span.
Ribosomal RNA (rRNA) : rRNA constitutes redundant nature upto 80% of total RNA of the cell. It occurs in ribosomes, which are nucleoprotein molecules.
Inside the ribosomes of eukaryotic cells rRNA occurs in the form of the particles of four different dimensions. These are designated 28S, 18S, 5.8S and 5S.
The 28S and 5S molecules occur in large subunit (60S subunit) of ribosome, whereas 18S molecules is present in the small subunit (40S subunit) of ribosome. In prokaryotic cells there are only 23S, 16S and 5S rRNA are found. Which are synthesized in Nucleolus / SAT region.
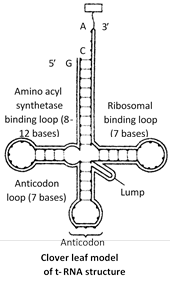
Transfer RNA (tRNA) : The transfer RNA is a family of about 60 small sized ribonucleic acids which can recognize the codons of mRNA and exhibit high affinity for 21 activated amino acids, combine with them and carry them to the site of protein synthesis. tRNA molecules have been variously termed as soluble RNA or supernatant RNA or adapter RNA. It is about 0-15% of RNA of the cell.
tRNA molecules are smallest, containing 75 to 80 nucleotides. The 3¢ end of the polynucleotide chain ends in CCA base sequence. This represents site for the attachment of activated amino acid. The end of the chain terminates with guanine base. The bent in the chain of each tRNA molecule contains a definite sequence of three nitrogenous bases, which constitute the anticodon. It recognizes the codon on mRNA.
Most accepted model for t-RNA structure is clover leaf model, which way given by Robert Holley (1965) along with H.G. Khorana and Nirenberg (for yeast alanyl t-RNA) and for this work, they were awarded Nobel prize in 1968.
Four different region or special sites can be recognised in the molecule of tRNA.These are :
Amino acid attachment site : It occurs at the 3¢ end of tRNA chain and has OH group combines with specific amino acid in the presence of ATP forming amino acyl tRNA.
Site for activating enzymes : Dihydrouridine or DHU loop dictate activation of enzymes.
Anticodon or codon recognition site : This site has three unpaired bases (triplet of base) whose sequence is complementary with a codon in mRNA.
Ribosome recognition site \[({{T}_{\Psi }}C):\]This helps in the attachment of tRNA to the ribosome.
Other types of RNA
Small nuclear RNA (snRNA) : It is a small sized RNA present in the nucleus. SnRNA takes part in splicing (U1 and U2), rRNA processing (U3) and mRNA processing.
Small cytoplasmic RNA (scRNA) : It is small sized RNA occurring free in the cytoplasm. It helps in taking and binding a ribosome to endoplasmic reticulum for producing secretory proteins.
You need to login to perform this action.
You will be redirected in
3 sec
