Notes - Classification and Factors Affecting Enzyme
Category : NEET
Classification and factors affecting enzyme
Nomenclature and Classification.
Dauclax, (1883) introduced the nomenclature of enzyme. Usually enzyme names end in suffix-ase to the name of substrate e.g. Lactase acts on lactose, maltase act on maltose, amylase on amylose, sucrase on sucrose, protease on proteins, lipase on lipids and cellulase on cellulose. Sometimes arbitrary names are also popular e.g. Pepsin, Trypsin and Ptylin etc. Few names have been assigned as the basis of the source from which they are extracted e.g. Papain from papaya, bromelain from pineapple (family Bromeliaceae). Enzymes can also be named by adding suffix?ase to the nature of chemical reaction also e.g. oxidase, dehydrogenase, catalase, DNA polymerase.
(1) According to order classification
The older classification of enzymes is based on the basis of reactions which they catalyse. Many earlier authors have classified enzymes into two groups:
(i) The hydrolysing enzymes
(ii) The desmolysing enzymes.
Other classify enzymes into three groups
(i) The hydrolysing enzymes
(ii) The transferring enzymes
(iii) The desmolysing enzymes
In the first classification transferring enzymes are included in the hydrolysing enzymes since some of them are known to act as transferring as well as hydrolysing enzymes.
(i) Hydrolysing enzyme: The hydrolysing enzymes of hydrolases catalyse reactions in which complex organic compounds are broken into simpler compounds with the addition of water. Most of the hydrolysing (digestive) enzymes are located in lysosomes. Depending upon the substrate hydrolysing enzymes are:
(a) Carbohydrases : Most of the polysaccharides, disccharides or small oligosaccharides are hydrolysed to simpler compounds, e.g., hexoses or pentoses under the influence of these enzymes.
Lactase on lactose to form glucose to galactose, sucrase/invertase on surcose to form glucose and fructose, amalyse or diastase on starch to form maltose, maltase on maltose to form glucose, cellulase on cellulose to produce glucose.
(b) Easterases : These enzymes catalyse the hydrolysis of substances containing easter linkage, e.g., fat, pectin, etc. into an alcoholic and an acidic compound.
\[\text{Fat}\xrightarrow{\text{lipase}}\text{Glycerol }+\text{ Fatty acid}\]
\[\text{Phosphoric acid easters }\xrightarrow{\text{phosphatase }}\text{Phosphoric acid }+\text{ Other compounds }\]
(c) Proteolytic enzymes : The hydrolysis of proteins into peptones, polypeptides and amino acids is catalysed by these enzymes
\[\text{Protein}\xrightarrow{\text{Pepsin}}\text{Peptones}\]
\[\text{Polypeptides}\xrightarrow{\text{Peptidases}}\text{Amino acids}\]
(d) Amidases : They hydrolyse amides into ammonia and acids.
\[\text{Urea}\xrightarrow{\text{Urease}}\text{Ammonia}+\text{Carbon dioxide}\]
\[\text{Asparagine}\xrightarrow{\text{asparaginase}}\text{Ammonia}+\text{Aspartic acid}\]
(ii) Desmolysing enzymes : Most of the desmolysing enzymes are the enzymes of respiration e.g. oxidases, dehydrogenases, (concerned with transfer of electrons), transaminases carboxylases etc.
(2) According to IUB system to classification
The number of enzymes is very large and there is much confusion in naming them. In 1961 the Commission on enzymes set up by the 'International Union of Biochemistry' (IUB) framed certain rules of their nomenclature and classification.
According to IUB system of classification the major points are :
· Reactions (and enzymes catalyzing them) are divided into 6 major classes each with 4-13 subclasses.
· The enzyme name has two parts-first name is of substrate. The second ending in ase indicates type of reaction.
· The enzyme has a systematic code No. (E.C.). The first digit denotes the class, the second sub-class, the third sub-sub-class and the fourth one is for the particular enzyme name. Thus, E.C. 2.7.1.1 denotes class 2 (Transferases)-subclass 7 (transfer of phosphate) sub-sub-class 1 (an alcohol functions as phosphate acceptor). The 4th digit indicates hexokinase.
Major classes of enzymes are as follows :
(i) Oxidoreductases : These enzyme catalyse oxidation reduction reactions, usually involving the transfer of hydrogen atoms or ions from one molecule to another. There are three main types of these enzymes :
(a) Oxidases : Where the hydrogen is transferred from a molecule to oxygen, e.g., cytochrome oxidase. They play very important role in E.T.S. in photosynthesis as well as respiration,
(b) Dehydrogenases : Where the hydrogen is transferred to a coenzyme such as NAD+, e.g, Succinic dehydrogenase. They help in oxidation of organic molecules during aerobic respiration.
(c) Reductase : It is cause addition of hydrogen or an electron and remove oxygen. e.g., Nitrate reductase requires NAD (coenzyme I) as coenzyme for the reaction.
(ii) Transferases : These enzyme catalyse the transfer of a specific group (e.g. amino, methyl, acyl, phosphate) from one kind of molecule to another e.g. transphosphorylases, transaminases, transpeptidases, transmethylases, kinases, etc.
(iii) Hydrolases : These enzyme catalyse the hydrolysis of organic foods i.e. the breakdown of large molecules by addition of water e.g.all digestive enzymes such as lipases (digest the stored food material of caster seeds) amylases, esterases, phosphatases, carbohydrases, proteases.
(iv) Lyases : These enzymes catalyses the breakage of specific covalent bonds and removal of groups without hydrolysis e.g. fumerases, carboxylases, aminases, histidine decarboxylase that splits C?C-bond of histidine, forming CO2 and histamine.
(v) Isomerases : These enzymes catalyses the rearrangement molecular structure to form isomers. e.g. phosphohexose isomerase (phosphoglucomutase) act on glucose 6-phosphate to form fructose 6-phosphate (both C6 compounds); epimerase.
(vi) Ligases or synthetases : These enzymes form bonds join two molecules together, using energy supplied from the breakdown of ATP,e.g., DNA ligase is used to repair breaks in DNA molecules. Amino-Acyl synthetase is used to activate t-RNA by attaching amino acid at 31 end. Tryptophase synthetase is used to convert tryptophase amino acid to IAA, etc.
Site of enzyme action.
All enzymes are produced in the living cells. About 3,000 enzymes have recorded. These are of two types with regard to the site where they act : intracellular and extracellular.
(1) Intracellular enzymes : Most of the enzymes remain and function inside the cells, They are called the intracellular enzymes or endoenzymes. Some of these enzymes are found in cytoplasmic matrix. Certain enzymes are bound to ribosomes, mitochondria and chloroplast etc.
(2) Extracellular enzymes : Certain enzymes leave the cells and function outside them. They are called the extracellular enzymes or exoenzymes. They mainly include the digestive enzymes. e.g. salivary amylase, gastric pepsin, pancreatic lipase secreted by the cells of salivary glands, gastric glands and pancreas respectively, lysozyme present in tears and nasal secretion.
Rennet tablets with enzyme renin from calf's stomach are widely used to coagulate protein caseinogen for cheese (casein) formation.
Mechanism of enzyme action.
Chemical reaction takes place between molecules when they are activated. An activated molecule is at a higher energy level than other molecules. Increase in the number of activated molecules increases the speed of the chemical reaction. Energy is required to bring the inert molecules into the activated state. The amount of energy required to raise the energy of molecules at which chemical reaction can occur is called activation energy. Enzymes act by decreasing the activation energy so that the number of activated molecules is increased at lower energy levels. If the activation energy required for the formation of the enzyme-substrate complex is low, many more molecules can participate in the reaction than would be the case if the enzyme were absent.
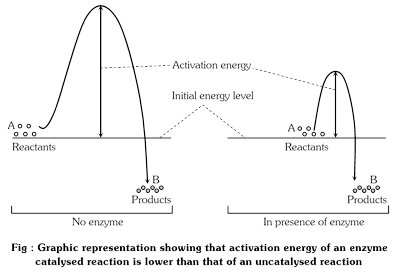
For example, activation energy, without adding the enzyme for the conversion of H2O2 into H2O and O2 is 18,000 calories per mole. But after addition of enzyme (catalase) the value is reduced to only 5,500 calories.
\[{{H}_{2}}{{O}_{2}}\underset{(\text{an}\,\text{enzyme})}{\mathop{\xrightarrow{catalase}}}\,2{{H}_{2}}O+{{O}_{2}}\]
Mode of enzyme action
In 1913 Michaelis and Menten proposed that for a catalylic reaction to occur it is necessary that enzyme and substrate bind together to form an enzyme substrate complex.
It is, however, difficult to demonstrate such complexes experimentally. Subsequently, the complex breaks up releasing the product and regenerating the original enzyme molecules for reuse.
\[\underset{\text{(Enzyme)}}{\mathop{\text{E}}}\,+\underset{(\text{Substrate)}}{\mathop{\text{S}}}\,\to \underset{(\text{Enzyme-substrate Complex)}}{\mathop{\text{E-S Complex}}}\,\]
\[\text{E-S Complex}\to \underset{\text{(Enzyme)}}{\mathop{\text{E}}}\,+\underset{(\text{Product)}}{\mathop{\text{P}}}\,\]
It is amazing that the enzyme-substrate complex breaks up into chemical products different from those which participated in its formation (i.e., substrates).
On the surface of each enzyme there are many specific sites for binding substrate molecules called active sites or catalytic sites. Structurally, each active site is an indentation on enzyme surface. It is lined by approximately 20 amino acids. During the course of reaction the substrate molecules occupy these sites. The active sites are located close to each other, hence, the substrate molecules also come close and react with one another. It is thought that when enzyme and substrates bind together, the shape of the enzyme molecule undergoes slight change. This produces strain in critical bonds in the substrate molecules and as a result these bonds break and new bonds are formed. The new chemical compound thus formed has little affinity for the enzyme and moves away from it.
There are two views regarding the mode of enzyme action :
(1) Lock and key hypothesis
(2) Induced fit hypothesis
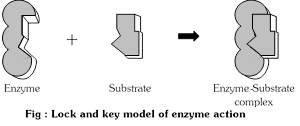
(1) Lock and key hypothesis : The hypothesis was put forward by Emil Fisher (1894) . According to this hypothesis the enzyme and its substrate have a complementary shape. The specific substrate molecules are bound to a specific site of the enzyme molecule.
The theory can be explained easily by the fact that a particular lock can be opened by a particular key specially designed to open it. Similarly enzymes have specific sites where a particular substrate can only be attached. The lock and key model accounts for enzyme specificity.
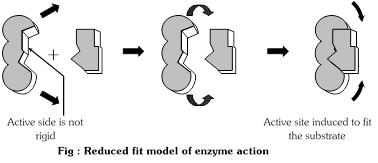
(2) Induced fit hypothesis : This hypothesis was proposed by Daniel, E. Koshland (1959).
According to this view, the active sites of an enzyme are not rigid. When the substrate binds to enzyme, it may induce a change in shape of the enzyme molecule in such a way that it is fit for the substrate-enzyme interaction. The change in shape of the enzyme molecules can put strain on the substrate. This stress may help bonds to break, thus promoting the reaction. In other words:
According to this theory active site of the enzyme contains two group-buttressing and catalytic. The buttressing group is meant for supporting the substrate. The catalytic group is able to weaken the bonds of reactants by electrophilic and nucleophilic forces. Both buttressing and catalytic groups are normally at a distance. When substrate comes in contact with the buttressing group, the active site of enzyme undergoes conformational changes to bring the catalytic group opposite the substrate bonds to be broken.
 |
Properties of enzymes.
The common properties of enzymes are listed below :
(1) Molecular weight : Enzymatic proteins are substances of high molecular weight. Peroxidase one of the smaller enzymes has molecular weight of 40,000, where as catalase one of the largest-has a molecular weight of 250,000 (urease 483,000). Enzyme molecules are therefore larger than those of usual simple organic substances but are nevertheless small enough to dissolve completely in aqueous media to yield clear nonturbid solution.
(2) Amphoteric nature : Each molecule of enzyme possess numerous groups which yield H+ in slightly alkaline solutions and groups which yield OH? ions in slightly acidic solutions. Unlike many other substances, therefore, the enzymatic protein is amphoteric, i.e., capable of ionizing either as an acid or as a base depending upon the acidity of the external solution.
(3) Colloidal nature : All enzymes are colloidal in nature and thus provide large surface area for reaction to take place. They posses extremely low rates of diffusion and form colloidal system in water.
(4) Specificity of enzyme : Most of the enzymes are highly specific in their action. A single enzyme will generally catalyse only a single substrate or a group of closely related substrates. e.g. the enzyme lactase catalyzes the hydrolysis of lactose and no other disaccharide and the enzyme malic dehydrogenase removes hydrogen atom from malic acid and not from other keto acids. The enzymes posses active sites which are highly specific centres composed of varying number and sequence of amino acids. The active site possess a particular binding site which complexes only with specific substrate. Thus, only a suitable substrate fulfils the requirements of active site and closely fixes with it.
(5) Heat specificity : The enzymes are thermolabile i.e. heat sensitive. They function best at an optimum temperature (20°C-40°C). Their activity decrease with decrease as well as increase in temperature and stops at 0°C and above 80°C.
(6) Catalytic properties : Enzymes are active in extremely small amounts, e.g. on molecule of invertase can effectively hydrolyze 1,000,000 times its own weight of sucrose. One molecule of catalase is able to catalyze conversion of 5,000,000 molecules of hydrogen peroxide. The enzyme remains unchanged, qualitatively or quantitatively after the reaction.
![]() (7) Reversibility of reaction : The enzyme-controlled reactions are reversible. The enzymes affect only the rate of biochemical reactions, not the direction. They can accelerate the reaction in either direction, i.e. onwards and backwards depending upon the availability of suitable energy sources e.g. Lipase can catalyase splitting of fat into fatty acids and glycerol as well as synthesis of fatty acids and glycerol into fats.
(7) Reversibility of reaction : The enzyme-controlled reactions are reversible. The enzymes affect only the rate of biochemical reactions, not the direction. They can accelerate the reaction in either direction, i.e. onwards and backwards depending upon the availability of suitable energy sources e.g. Lipase can catalyase splitting of fat into fatty acids and glycerol as well as synthesis of fatty acids and glycerol into fats.
Fat Glycerol+ Fatty acid
(8) pH sensitivity : The enzymes show maximum activity at an optimum pH (6-7.5). Their activity slows with decrease and increase in pH till it stops. Each enzyme has its own different favourable pH value.
(9) High efficiency : The effectiveness of an enzymic reaction is expressed in terms of its turn over number or catalytic centre activity means number of substrate molecules on which one enzymes molecules acts in one minute.
Turn over number depends on the number of active sites of an enzyme. An active site is an area of the enzyme which is capable of attracting and holding particular substrate molecules by its specific charge, size and shape so as to allow the chemical change, Enzymes show 3-D structure. R (alkyl) groups of amino acids from active sites during folding polypeptide chains. Usually 3-12 amino acids form an active site. More the member of active sites, more is the turnover number of enzymes. Enzyme react with substrate only at these active sites. The whole surface of enzyme is not reactive. Enzymes have high turn over number (Catalytic number).
Highest turn over number is of carbonic anhydrase (36 million/min or 600000 per second) and lowest is of lysozymes (30/min or 0.5 per second). So carbonic anhydrase is fastest enzyme. It has zinc as activator. It hydrates 36 million CO2 molecules per minute in RBC into H2CO3.
Turn over number depends upon number of active sites, rapidity of reaction and separation of end product.
(10) Team work : The enzymes generally work in teams in the cell, the product of one enzyme controlled reaction serving as the substrate for the next. In germinating seeds, starch is changed into glucose by two enzymes : amylase and maltase. Amylase splits the starch into the double sugar maltose, which is then broken by maltase into the single sugar glucose. Eleven different enzymes work sequentially to convert glucose to lactic acid in animal as well as plant cells.
(11) Destruction by poisons : Enzymatic activity can be retarded or inhibited by the use of toxic substances like cyanide and iodoacetic acid, cyanide destroys the respiratory enzyme cytochrome oxidase.
Enzyme inhibition.
Interaction of an enzyme with substances other than the normal substrate changes the structure of enzyme. If this change occurs, there is loss in catalytic efficiency or complete in activation of enzyme. Inhibition may be of following types :
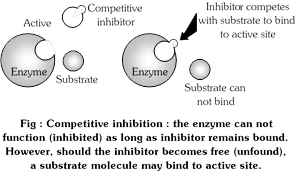
(1) Competitive inhibition : Substances (inhibitors) which are structurally similar to the substrates and complete for the active site of the enzyme are known as competitive inhibitors. Usually such inhibitors show a close structural resemblance to the substrates to the enzyme they inhibit. In such a case, inspite of enzyme substrate complex, enzyme inhibitor complex is formed and enzyme activity is inhibited.
 \[\underset{\text{Enzyme}}{\mathop{\text{E}}}\,+\underset{\text{inhibitor}}{\mathop{\text{I}}}\,\to \underset{\text{Enzyme}-\text{inhibitor}\,\text{complex(EI)}}{\mathop{\text{EI}}}\,\]
\[\underset{\text{Enzyme}}{\mathop{\text{E}}}\,+\underset{\text{inhibitor}}{\mathop{\text{I}}}\,\to \underset{\text{Enzyme}-\text{inhibitor}\,\text{complex(EI)}}{\mathop{\text{EI}}}\,\]
The concentration of EI complex depends on the concentration of free inhibitor. Because EI complex readily dissociates, the empty active sites are then available for substrate binding. The effect of a competitive inhibitor on activity is reversed by increasing the concentration of substrate.
A classic example of competitive inhibition is succinic acid dehydrogenase which oxidises succinic acid to fumaric acid. If concentration of malonic acid, is added, the activity of succinic dehydrogenase decreases rapidly. Hence malonic acid acts as a competitive inhibitor since it has structural resemblance to succinc acid.
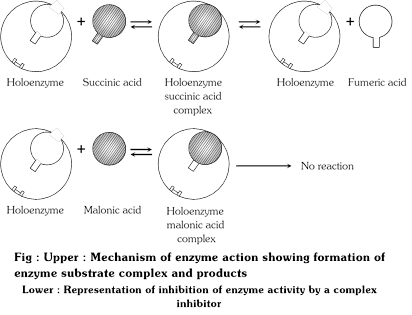 |
The competitive inhibition can be reversed by increasing the concentration of the substrate. Competitive inhibitors are used in control of bacterial pathogens. Sulpha drugs is similar to PABA (para aminobenzoic acid) act as competitive inhibitors in the synthesis of folic acid in the bacterial cells because they compete with p-amino benzoic acid for the active sites of the enzyme and check the synthesis.
(2) Non-competitive inhibition : These substances (poisons) do not combine with active sites but attach somewhere else and destroy the activity of enzyme.
Both EI and ES complexes are formed. Inhibitor binding alters the three dimensional configuration of the enzyme and thus blocks the reaction. Non competitive inhibitor do not compete directly with the substrate for binding to the enzyme.
The non-competitive inhibition can not be reversed by increasing the concentration of the substrate i.e. irreversible. e.g. cyanide inhibits the mitochondrial enzyme cytochrome oxidase which is essential for cellular respiration. This kills the animals. Cyanide (inhibitor) does not compete for active sites of enzyme with substrate because it has no similarity with substrate (cytochrome) but it acts at some other site of enzyme.
More AMP is a non competitive inhibitor of fructose biphosphate phosphatase, the enzyme that catalyses the conversion of fructose 1, 6 biphosphate to fructose 6 phosphate. Toxic metal ions destroy essential sulphydryl groups of certain enzymes.
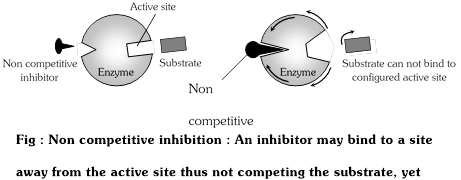 |
|
S. No. |
Competitive inhibition |
|
Non competitive inhibition |
|
(1) |
The structure of the inhibitor molecule is similar to the substrate. |
(1) |
The structure of the inhibitor is different from the substrate. |
|
(2) |
The inhibitor gets attached to the enzyme's active site. |
(2) |
The inhibitor forms a complex at a site other than the active site on the enzyme. |
|
(3) |
The reaction can be reversed at any stage by increasing the substrate concentration. |
(3) |
The reaction will keep on decreasing till there is saturation of inhibitor. |
|
(4) |
The substrate competes with the inhibitor for the position of the active site. |
(4) |
The substrate does not compete with the inhibitor as the name indicates. |
|
(5) |
The inhibitor does not alter the structure of the enzyme. |
(5) |
The inhibitor alters the structure of the enzyme in such a way that even if the substrate gets attached, the end products will not be formed. |
|
(6) |
The competition of pesticides with the neurotransmitter chemicals while binding to chemoreceptor sites on dendrites. |
(6) |
Cyanide and azides combines with the prosthestic groups of cytochrome oxidase and inhibits the electron transport chain. |
![]() (3) Feedback inhibition : In number of cases, accumulation of the final product of the reaction is capable of inhibiting the first step of reaction.
(3) Feedback inhibition : In number of cases, accumulation of the final product of the reaction is capable of inhibiting the first step of reaction.
\[A\xrightarrow{{{E}_{1}}}B\xrightarrow{{{E}_{2}}}C\xrightarrow{{{E}_{3}}}D\xrightarrow{{{E}_{4}}}P\]
The product P checks the activity of enzyme which converts A into B. It is quite useful mechanism because it checks the accumulation of products.
The phenomenon in which the end product of a metabolic pathway can regulate its own production by inhibition of the sort is called feed back inhibition or negative feed back inhibition. This type of inhibition can be shown in Escherichia coli bacterium which synthesises the amino acid isoleucine from a substrate threonine by a series of intermediate reactions (i.e. \[\alpha \]ketobutyrate threonine deaminase, \[\alpha \]Aceto hydroxy butyrate, \[\alpha \]keto\[\beta \]methyl valerate etc).
 |
When isoleucine accumulates in amounts more than required, it stops its own production by inhibiting the activity of the enzyme. Threonine deaminase which catalyzes the first reaction of the series. This type of metabolic control in which the first enzyme of a series is inhibited by the end product, is known as end product inhibition.
 (4) Allosteric inhibition (modulation) : Allosteric literally means ' another place'. Still other inhibitors join an enzyme at a specific site and change the form of the active site meant for the substrate. These inhibitors are known as modifiers or modulators and the sites where they fit in are called allosteric sites. Modulators are of two types-positive (activators) and negative (inhibitors). Allosteric enzyme phosphofructokinase is activated by ADP and inhibited by ATP. Diphosphofructose, phosphate is activated by ATP and inhibited by AMP. Change of active site form prevent the binding of substrate to the enzyme and stops the reaction. The process is called allostery or allosteric inhibition, The enzyme with allosteric sites are called allosteric enzymes. Jacob and Monod have termed this phenomenon as allosteric transition.
(4) Allosteric inhibition (modulation) : Allosteric literally means ' another place'. Still other inhibitors join an enzyme at a specific site and change the form of the active site meant for the substrate. These inhibitors are known as modifiers or modulators and the sites where they fit in are called allosteric sites. Modulators are of two types-positive (activators) and negative (inhibitors). Allosteric enzyme phosphofructokinase is activated by ADP and inhibited by ATP. Diphosphofructose, phosphate is activated by ATP and inhibited by AMP. Change of active site form prevent the binding of substrate to the enzyme and stops the reaction. The process is called allostery or allosteric inhibition, The enzyme with allosteric sites are called allosteric enzymes. Jacob and Monod have termed this phenomenon as allosteric transition.
![]() An example of allosteric enzyme inhibition is hexokinase that converts glucose to glucose 6-phosphate. Glucose 6-phosphate causes allosteric inhibition of hexokinase. This is called feedback allosteric inhibition.
An example of allosteric enzyme inhibition is hexokinase that converts glucose to glucose 6-phosphate. Glucose 6-phosphate causes allosteric inhibition of hexokinase. This is called feedback allosteric inhibition.
\[\text{Glucose}+\text{ATP}\xrightarrow{\text{Hexokinase}}\text{Glucose 6-phosphate }+\text{ADP}\]
Some terms regarding Enzymes .
(1) Zymogen : Certain enzymes are produced by the living cells in an inactive (non-functional) form. They are called the zymogens or proenzymes. It is then converted, usually by proteolysis (hydrolysis of the protein), to the active form when it has reached the site of its activity. Pepsinogen and trypsinogen are zymogens produced by gastric glands and pancreas respectively. They are necessary to life because they degrade dietary proteins into amino acids that are used by the cell.
Pepsinogen is changed to active pepsin by hydrogen ions in the stomach. Trypsinogen is activated to trypsin by an enzyme enterokinase in the small intestine. Once small amount of pepsin or trypsin is formed, it itself catalyzes the activation of remaining proenzyme. This process is called autocatalytic reaction, or autocatalysis.
(2) Isoenzyme (Isozymes) : There are certain enzymes which have slightly different molecular structure but performing the same catalytic function. Such enzymes are called isoenzymes or simply isozymes. Isoenzyme of an enzyme differ from one another in their amino acid sequence, molecular weight, immunological and electrophoretic behaviours. Hence, they can be separated by electrophoresis. When the variants of an enzyme are within the same species of an organism they are called intraspecific or ontogenic variants; when they are from different species they are called interspecific or phylogenetic variants. Isoenzymes provide a clue to the genetic relationships of organism. Similarity in isoenzymes is corrected with similarity in genotype.
The isoenzyme differ in optimum activity and enable the organism adapt to varied environmental conditions. It is held that the isoenzymes are produced by genetic changes during evolution.
Isoenzymes may be homologous or analogous. Homologous isoenzymes have essentially similar molecular structure and catalytic properties. Analogous isoenzymes, though catalyse similar reaction, have different molecular structure and catalytic properties.
More than 100 enzymes are known to have isoenzyme. A good example of isoenzyme is lactic dehydrogenase (LDH). It catalyzes change of pyruvate to lactate. There are five LDH isoenzymes in muscles of heart have been dilineated designated as\[LD{{H}_{1}}-LD{{H}_{5}}\]. The different LDH isoenzymes differs significantly in maximum activities \[\left( {{V}_{max}} \right)\] and in Michaelis constant \[\left( {{K}_{m}} \right)\]for their substrates especially for pyruvate. Alcohol dehydrogenase has four isoenzyme in maize. a-amylase (wheat endosperm) has 16 isoenzymes.
(3) Inducible enzyme : An enzyme which is synthesized only in the presence of its substrate (inducer) is called inducible enzyme e.g., \[\beta \]-galactosidase.
(4) Constitutive enzymes (House keeping enzyme) : The enzyme which are found in constant amounts under different growth conditions (regardless of its metabolic states) are called constitutive enzyme e.g. enzymes of sugar break down i.e. glycolysis.
(5) Repressible enzyme : The presence of a specific substance may inhibit continued production of specific enzyme (enzyme repressor) e.g. glucokinase.
(6) Ribozymes : Study of post transcriptional processing of RNA molecules has led to the most exciting discovery of the existence of some catalytic RNA molecules which have been called as RNA enzymes or ribozymes. All enzymes are not proteins as confirmed by Cech (1981) and Altman (1983). Ribozyme and RNAase-P are two non protein enzyme where RNA acts as catalyst. Ribozyme was reported from Tetrahymens (a protozoans) by Cech. The substrate for ribozyme is usually an RNA molecule. RNAase-P (Ribonuclease) discovered by Altman.
(7) Michaelis constant : Michaelis and Menten (1913) introduced a constant Km (Michaelis constant).
It is a mathematical derivative or constant which indicates the substrate concentration at which the chemical reaction catalysed by an enzyme attains half its maximum velocity\[\left( {{V}_{max}} \right)\].
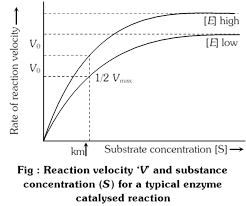 \[{{K}_{m}}\] indicates affinity of the enzyme for its substrate.
\[{{K}_{m}}\] indicates affinity of the enzyme for its substrate.
\[{{K}_{m}}=\frac{1}{2}{{V}_{\max }}\]
\[{{K}_{m}}\] value differs from substrate to substrate because different enzymes differ in their affinity towards different substrate. A high \[{{K}_{m}}\]indicates low affinity while a low \[{{K}_{m}}\]shows strong affinity. Protease acts on different proteins. So it \[{{K}_{m}}\] value will differ from protein to protein.
They propose general theory of enzyme action and kinetics. According to them it accounts for most of the features of enzyme catalyzed reactions. Enzyme combine with substrate to form enzyme-substrate (ES) complex and subsequently breaks down to product, regenerating the free enzyme.
![]()
![]()
Where S is the substrate; E is the enzyme; ES is the enzyme-substrate;
![]()
are rate constant.
The Michaelis Menten equation description how reaction relatively varies with substrate concentration as given
V0 =
![]()
Where \[{{V}_{0}}\]is the initial reaction; \[{{V}_{max}}\]is the maximum relative or the reaction rate with excess substrate; \[{{K}_{m}}\]is the Michaelis constant\[={{K}_{2}}+{{K}_{3}}/{{K}_{1}};\text{ }\left[ S \right]\]is the substrate concentration.
The above reaction show that the greater the affinity between an enzyme and its substrate, the lower the\[{{K}_{m}}\] (in units moles per litre) of the enzyme substrate reaction. Stated inversely, \[1/{{K}_{m}}~1/{{K}_{m}}\]is the measure of affinity of the enzyme for its substrate.
Allosteric enzymes do not show typical Michaelis Menten constant or allosteric enzymes do not obey \[{{K}_{m}}\]constant.\[{{K}_{m}}\] mostly lies between \[{{10}^{-1}}to\text{ }{{10}^{-6}}M\].
Factors affecting the Enzyme activity.
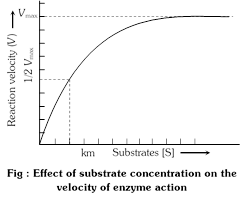 Like all chemical reactions, enzymatic reactions are sensitive to environmental conditions. Thus, the substrate concentration, enzyme concentration, pH. temperature and inhibitors all affect the rate of enzymatic reaction. These factors affect the active site of the enzyme and formation of the enzyme-substrate complex.
Like all chemical reactions, enzymatic reactions are sensitive to environmental conditions. Thus, the substrate concentration, enzyme concentration, pH. temperature and inhibitors all affect the rate of enzymatic reaction. These factors affect the active site of the enzyme and formation of the enzyme-substrate complex.
 (1) Substrate concentration : If there are more enzyme molecules than substrate molecules, a progressive increase in the substrate molecules increases the velocity of their conversion to products. However, eventually the rate of reaction reaches the maximum. At this stage the active sites of all the available enzyme molecules are occupied by the substrate molecules. Therefore, the substrate molecules occupy the active sites vacated by the products and cannot increase the rate of reaction further.
(1) Substrate concentration : If there are more enzyme molecules than substrate molecules, a progressive increase in the substrate molecules increases the velocity of their conversion to products. However, eventually the rate of reaction reaches the maximum. At this stage the active sites of all the available enzyme molecules are occupied by the substrate molecules. Therefore, the substrate molecules occupy the active sites vacated by the products and cannot increase the rate of reaction further.
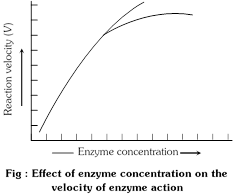
(2) Enzyme concentration : The rate of reaction is directly proportional to enzyme concentration. An increase in enzyme concentration will cause a rise in the rate of reaction up to a point and them the rate of reaction will be constant. Increasing the enzyme concentration increases the number of available active sites.
 (3) Product concentration : Accumulation of the product of enzyme reaction lowers the enzyme activity. Enzyme molecules must be freed to combine with more substrate molecules. Normally the product are quickly removed from the site of formation and the reaction does not suffer.
(3) Product concentration : Accumulation of the product of enzyme reaction lowers the enzyme activity. Enzyme molecules must be freed to combine with more substrate molecules. Normally the product are quickly removed from the site of formation and the reaction does not suffer.
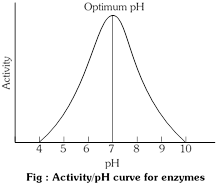
 (4) Hydrogen ion concentration (pH) : Some enzyme act best in an acid medium, other in an alkline medium, for every enzyme there is an optimum pH where its action is maximum e.g. 2 for pepsin, 6.8 for salivary amylase, 8.5 for trypsin. Most enzyme show maximum activity in a pH range of about 6.0 to 7.5 i.e.,near neutral pH (endoenzymes). A shift to the alkaline or acid side rapidly decreases the enzyme activity and finally stops it altogether. This is due to denaturation of enzyme molecule i.e. change in its physical structure. The H+ ions combines with negatively charged R groups on the enzyme. This electrically neutralizes the R groups and distrupt ionic bonds in the enzyme's folding pattern, thus changing its shape.
(4) Hydrogen ion concentration (pH) : Some enzyme act best in an acid medium, other in an alkline medium, for every enzyme there is an optimum pH where its action is maximum e.g. 2 for pepsin, 6.8 for salivary amylase, 8.5 for trypsin. Most enzyme show maximum activity in a pH range of about 6.0 to 7.5 i.e.,near neutral pH (endoenzymes). A shift to the alkaline or acid side rapidly decreases the enzyme activity and finally stops it altogether. This is due to denaturation of enzyme molecule i.e. change in its physical structure. The H+ ions combines with negatively charged R groups on the enzyme. This electrically neutralizes the R groups and distrupt ionic bonds in the enzyme's folding pattern, thus changing its shape.
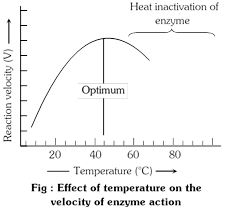
(5) Temperature : Within certain limits \[(5-40{}^\circ C)\] the rate of an enzyme catalysed reaction increases as the temperature increases. The \[{{Q}_{10}}\]of most enzymatic reactions is 2, i.e., every \[10{}^\circ C\]rise in temperature doubles the rate of reaction. Most enzymes show maximum activity in a temperature range of\[25\text{ }to\text{ }40{}^\circ C\]. Beyond this temperature, there is sharp fall in the rate of reaction.
Rise in temperature increases the kinetic energy of the molecules. Therefore, at higher temperature an increasing number of molecules have the required activation energy and can take part in chemical reactions. At higher temperatures, the kinetic activity of molecules in an enzyme becomes strong enough to break the weak hydrogen bonds that maintain the tertiary structure of the enzyme. Modification in the physical form of the enzyme results in the loss of its catalytic activity. This change in structure is called denaturation of protein. This is the permanent change, and the denatured enzyme protein remains inactive even if the temperature is then brought down. The enzymes are not destroyed by freezing, and regain their lost activity if the temperature is raised to normal.
Dry seeds can tolerate higher temperatures as compared to germinating seeds because dry seeds have dehydrated enzymes.
Deep freezing of food for preserving them for long periods is done not only to prevent the growth and multiplication of microorganisms but also to inactivate enzymes. It makes impossible for the microorganisms to digest the food.
(6) Enzyme inhibitors : Certain chemical compounds inhibit activity of enzyme molecules either permanently or temporarily. Thus, di-isopropyl flurophosphate (DFP) inhibits the action of various enzymes catalysing hydrolysis of ester linkage. Inhibition is permanent or irreversible. On the other hand, some antibacterial drugs and poisons do not cause permanent damage to the functional groups of the enzyme and therefore, if these (inhibitiors) are removed, the enzyme becomes fully functional.
(7) Poisons and radiation : Poisons such as cyanide and radiation destroy the tertiary structure of the enzymes, making them ineffective.
Important Tips
F Enzyme is called 'biological middle man'.
F Most of the vitamins of B complex group act as coenzyme.
F Smallest enzyme is peroxidase and largest being catalase found in perxisome.
F Myosin a structural component of muscle. It has ATPase activity also.
F Ki is dissociation constant of enzyme-inhibitor complex. It is applicable to competitive inhibitors only. Low Ki is essential for enzyme activity while a high Ki decreases it.
F Enzymes show reversible reactions and act by lowering energy of activation by more than 50%. They work in milliseconds and rate of enzyme to substrate is as high as 1:1000000.
F Synthesis of enzymes occur in polysome (aggregation of ribosomes).
F The structure of allosteric enzyme was studied by Monod et al (1965).
F Competitive inhibitor increase Michaelis constant (Km) but it has no effect on Vmax.
F Deficiency of an enzyme is called enzymophenia.
F cAMP mediated cascade model of enzyme regulation was proposed by Sutherland.
F All coenzymes are cofactors but all cofactors are not coenzymes.
F Allozyme are similar enzymes formed by different genes.
F Non competitive inhibitors decrease the \[{{V}_{max}}\]of enzyme but they have no effect on \[{{k}_{m}}\].
F Peptidases enzyme digests other enzymes.
F Viruses completely lack enzymes.
F Tertiory structure of protein component of enzyme is destroyed by a number of factors like heat, high energy radiation and salts of heavy metals \[\left( e.g.\text{ }A{{g}^{+}},\text{ }H{{g}^{2+}},\text{ }A{{s}^{+}}. \right)\]
F Markers are used for identification, Mitochondrial markers include succinate dehydrogenase, glutamate dehydrogenase and cytochrome oxidase. Glucose 6-phosphate is marker of E.R., RNA for ribosome, acid phosphatase for lysozyme, etc.
F Regulators of metabolism are enzymes, vitamins and hormones.
F RNA polymerase enzyme form RNA from DNA and DNA polymerase is responsible for synthesis of DNA from DNA.
F Enzyme that catalyses the conversion of soluble proteins into insoluble ones, process is called enzyme coagulation.
F PEP carboxylase catalyses the \[{{C}_{4}}\]cycle of photosynthesis.
F Fritz Lipmann discovered coenzymes.
F It is more difficult to denaturate the enzyme substrate complex than isolated enzyme.
F Albinism is caused by the deficiency of tyrosinase.
F Iron porphyrin coenzyme or cofactor is cytochrome.
F Nitrogenase enzyme is inactivated by oxygen.
F Nitrogenase enzyme is responsible for the reduction of molecular nitrogen to the level of ammonia in legumenous root nodule.
F Nitrate reductase enzyme is responsible for the formation of \[N{{O}_{2}}\].
F Amylopsin acts upon polysaccharide in alkaline medium.
F Due to enzymatic transformations huge amount of starch is deposited in potato tubers.
You need to login to perform this action.
You will be redirected in
3 sec
